Methods
To achieve benchmarking results, several data sets were used.
Genotypic and phenotypic maize data consisting of 279 samples, 3093
variant sites, and 1 measured trait were utilized for the analysis of
variant call format (VCF) import, generalized linear model (GLM)
association, mixed linear model (MLM) association, and kinship
generation times (Flint-Garcia et al., 2005). To illustrate the
effectiveness of the fast association method, 100 simulated RNA
expression traits for the prior genotype data was used. Trait data was
generated using the makeExampleDESeqDataSet() function from the R
package DESeq2 (Love et al., 2014). A larger genotypic data set
consisting of 1,210 samples and 2,255,405 variant sites was also
utilized for large VCF import and kinship generation times. All
benchmarks were generated using the microbenchmark() function from the R
package microbenchmark (Mersmann, 2019).
All benchmarks sans large VCF import and kinship generation times
were evaluated 100 times and recorded on a workstation running 16 GB of
RAM and 4 cores on an Intel® CoreTM i5-6500 CPU with a clock speed of
3.20 GHz and. Large VCF import and kinship generation benchmarks were
evaluated 10 times and recorded on a workstation running 256 GB of RAM
and 12 cores on an Intel® Xeon® CPU E5-2643 v3 with a clock speed of
3.40GHz.
Feature comparisons
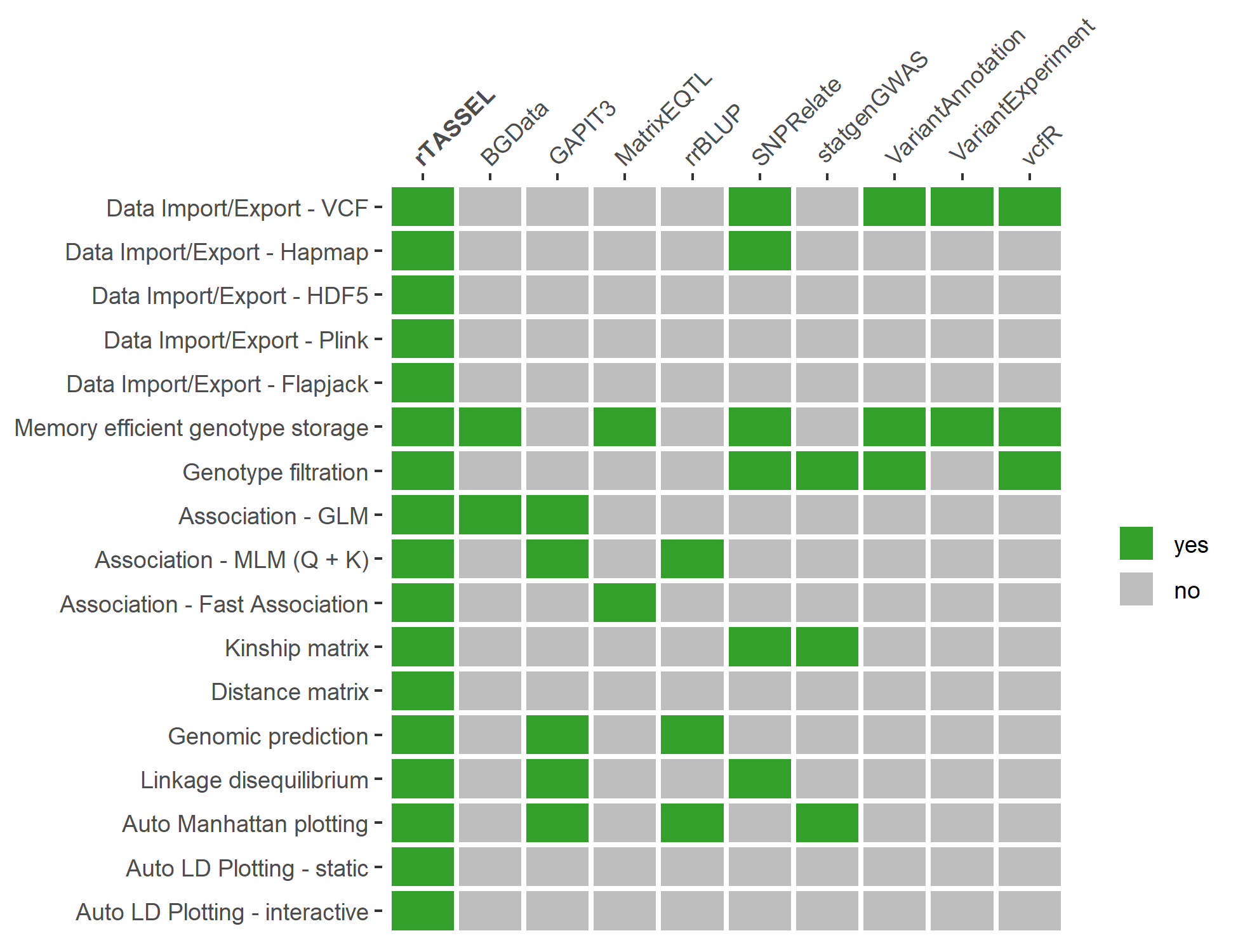
Feature comparisons of rTASSEL with
other R packages. Features of rTASSEL (y-axis) are compared
with other commonly-used R packages (x-axis). Packages that contain a
specified feature are highlighted green (yes) and grey (no) if they do
not contain a feature or are limited in scope. Association features for
packages are based on if said package contains methods for generalized
linear models, mixed linear models utilizing the “Q+K” method (Yu et
al., 2006), or multi trait fast association methods (Shabalin, 2012).
Kinship and distance matrix features denote if a package can return an n
x n matrix of values for further use. Packages that contain plotting
features indicate if the package contains an automated plot feature
instead of using base or grid-based R graphics (R Core Team, 2020) in
conjunction with data output. The packages used for this comparison are
BGData (Grueneberg and Campos, 2019), GAPIT3 (Wang and Zhang, 2020),
MatrixEQTL (Shabalin, 2012), rrBLUP (Endelman, 2011), SNPRelate (Zheng
et al., 2012), statgenGWAS (Rossum and Kruijer, 2020), VariantAnnotation
(Obenchain et al., 2014), VariantExperiment (Liu et al., 2020), and vcfR
(Knaus and Grünwald, 2017).
Speed comparisons
VCF import
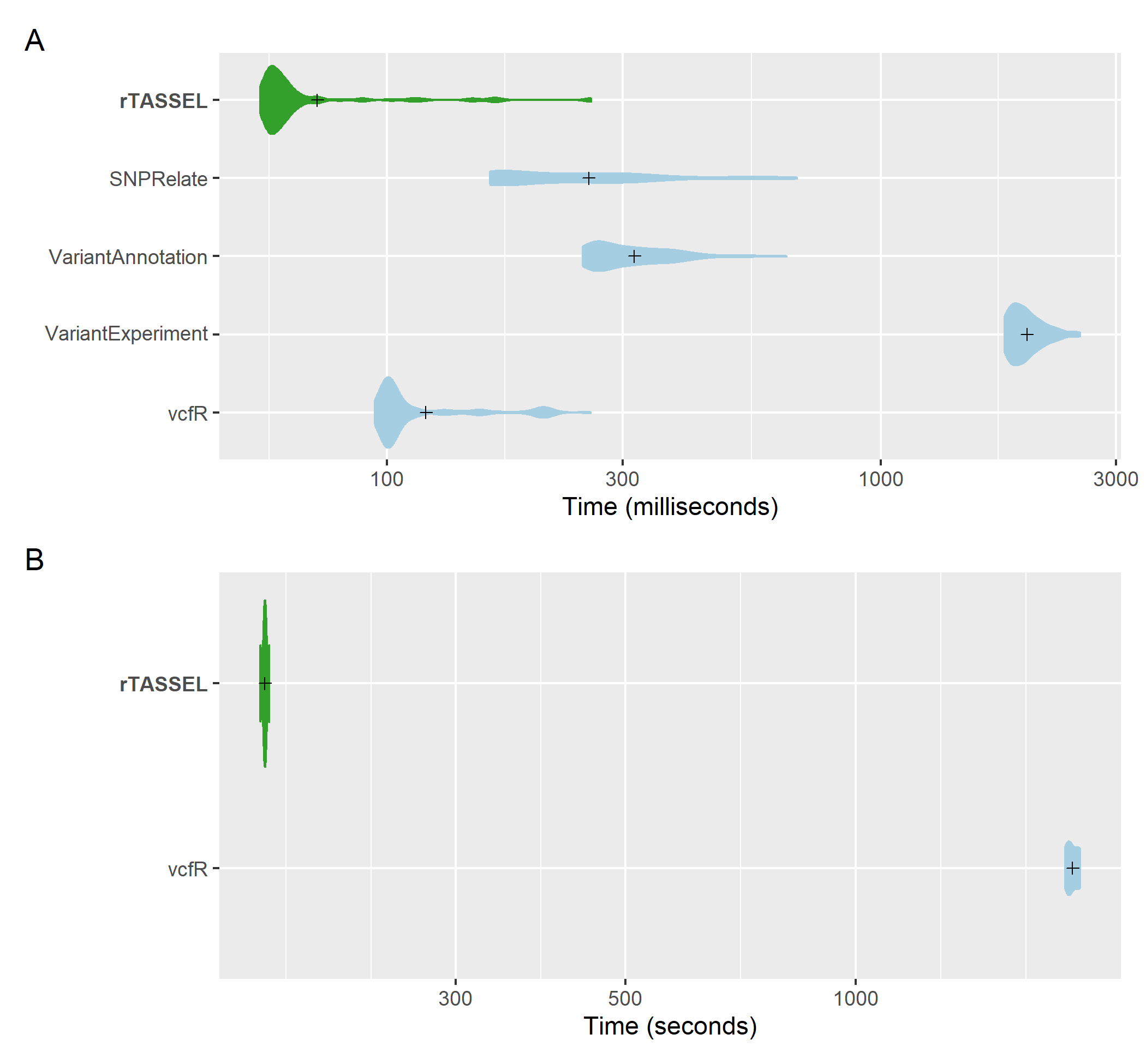
VCF import time comparisons of genotypic
data. A distribution of replicated benchmark evaluations with
recorded means (cross shapes) are plotted for rTASSEL and several R
packages: SNPRelate (Zheng et al., 2012), VariantAnnotation (Obenchain
et al., 2014), VariantExperiment (Liu et al., 2020), and vcfR (Knaus and
Grünwald, 2017). Import times are recorded for 279 samples x 3093
variant sites (A) and 1,210 samples x 2,255,405 variant sites (B).
Association methods
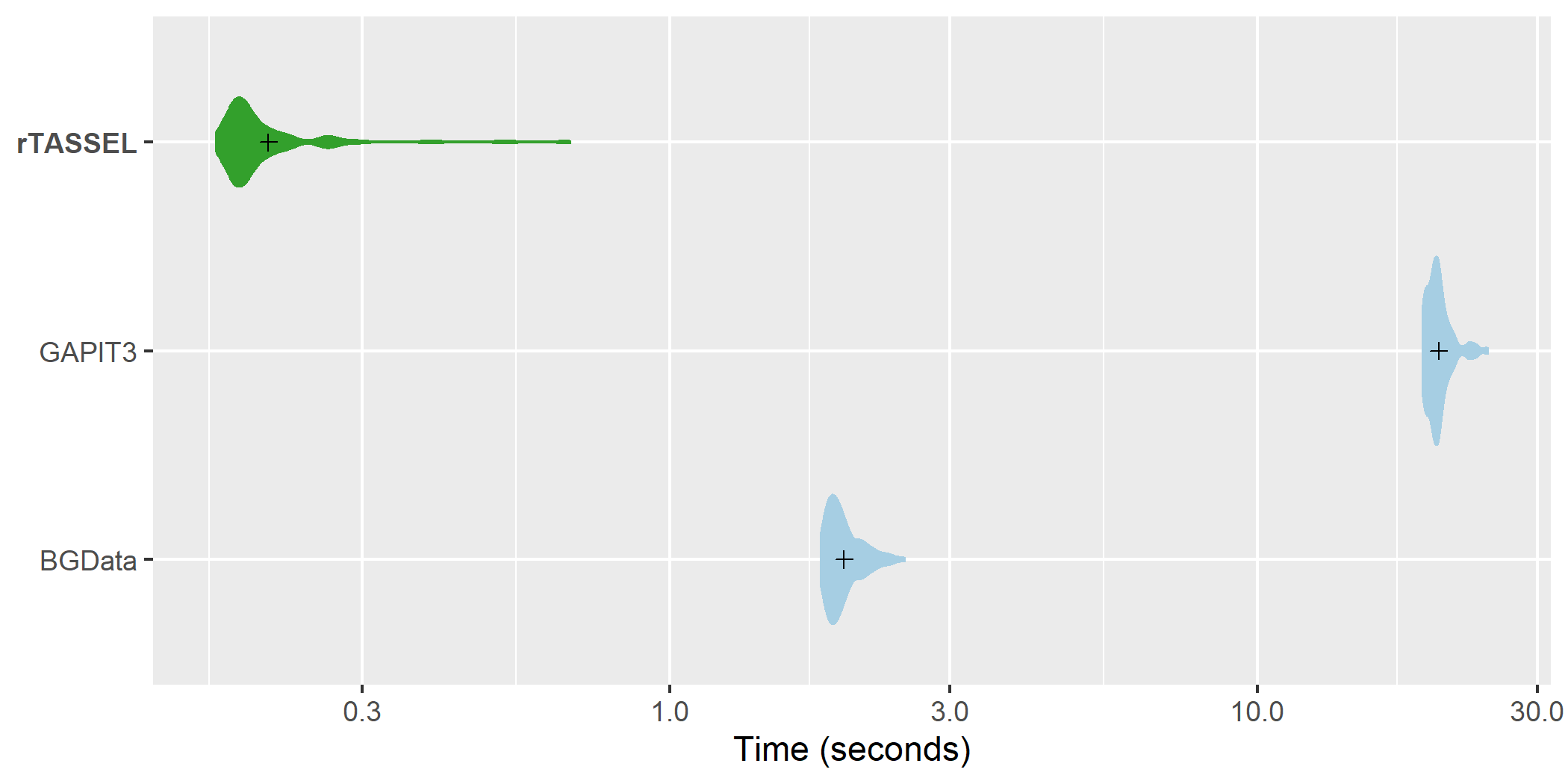
GLM association time
comparisons. A distribution of replicated benchmark evaluations
with recorded means (cross shapes) are plotted for rTASSEL and the R
packages GAPIT3 (Wang and Zhang, 2020) and BGData (Grueneberg and
Campos, 2019). Import times are recorded for 279 samples x 3093 variant
sites and 1 measured trait.
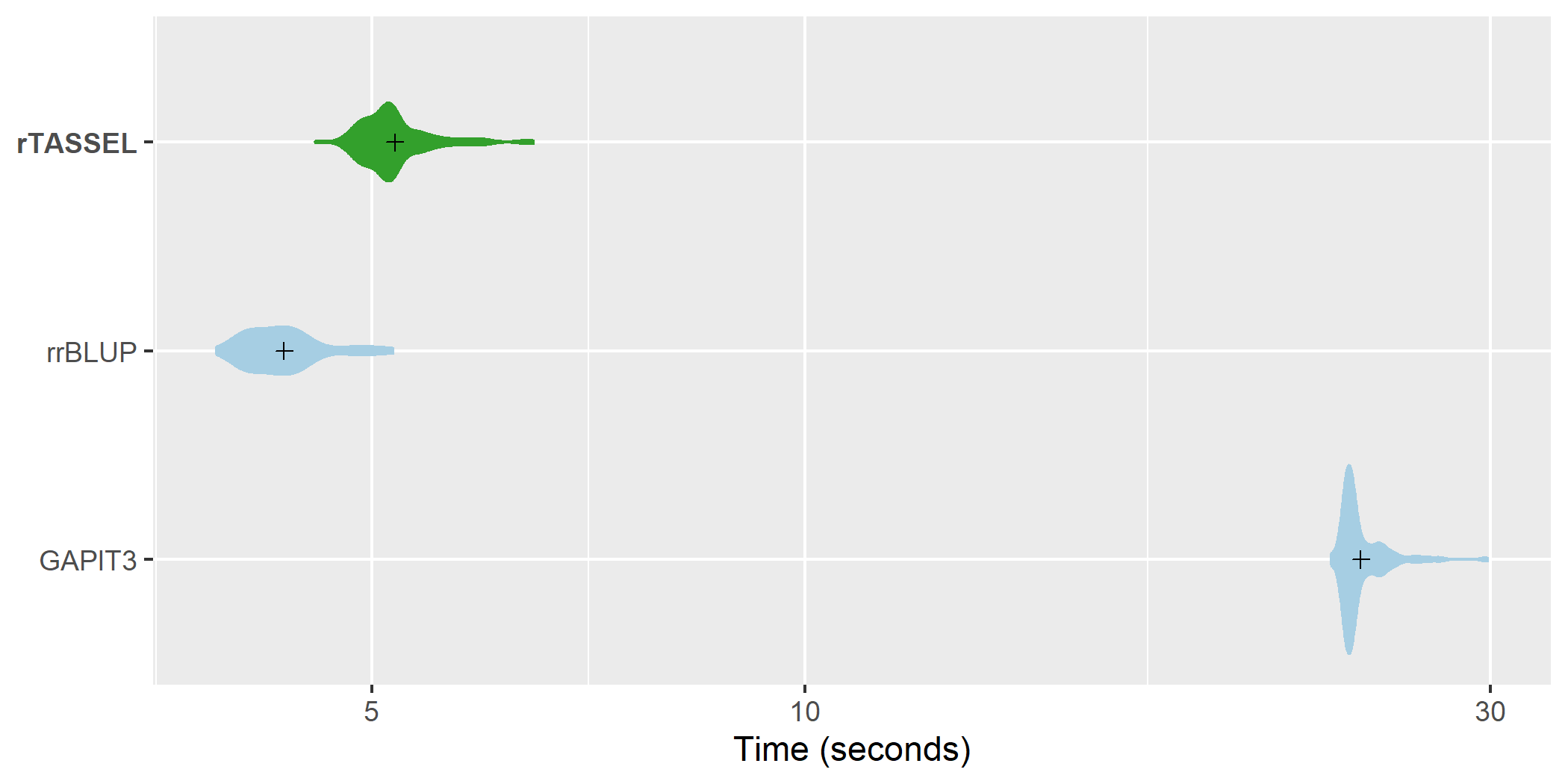
MLM association time
comparisons. A distribution of replicated benchmark evaluations
with recorded means (cross shapes) are plotted for rTASSEL and the R
packages rrBLUP (Endelman, 2011) and GAPIT3 (Wang and Zhang, 2020).
Import times are recorded for 279 samples x 3093 variant sites and 1
measured trait.
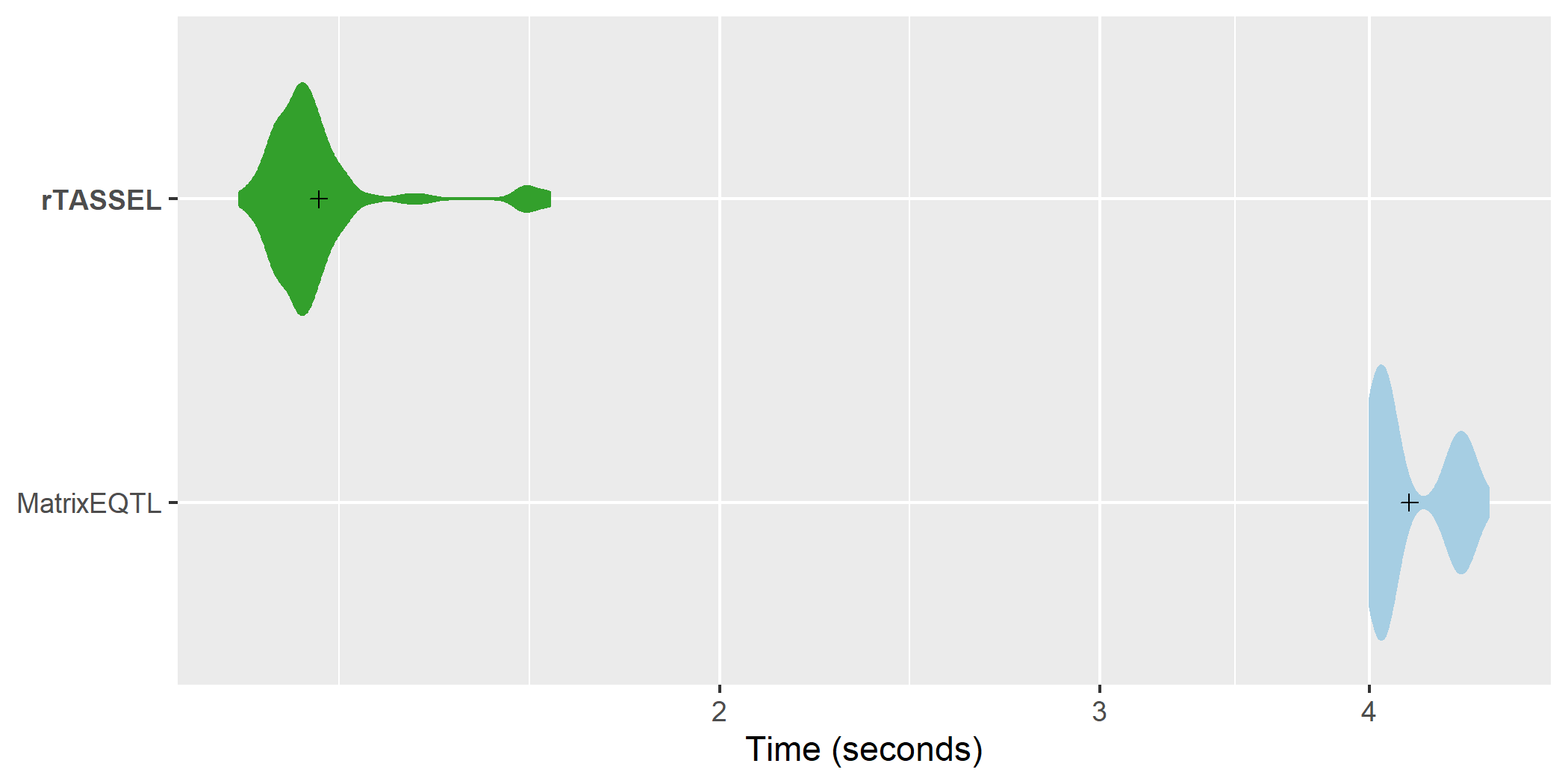
Fast association time
comparisons. A distribution of replicated benchmark evaluations
with recorded means (cross shapes) are plotted for rTASSEL and the R
package MatrixEQTL (Shabalin, 2012). Import times are recorded for 279
samples x 3093 variant sites and 100 simulated RNA expression
traits.
Kinship creation
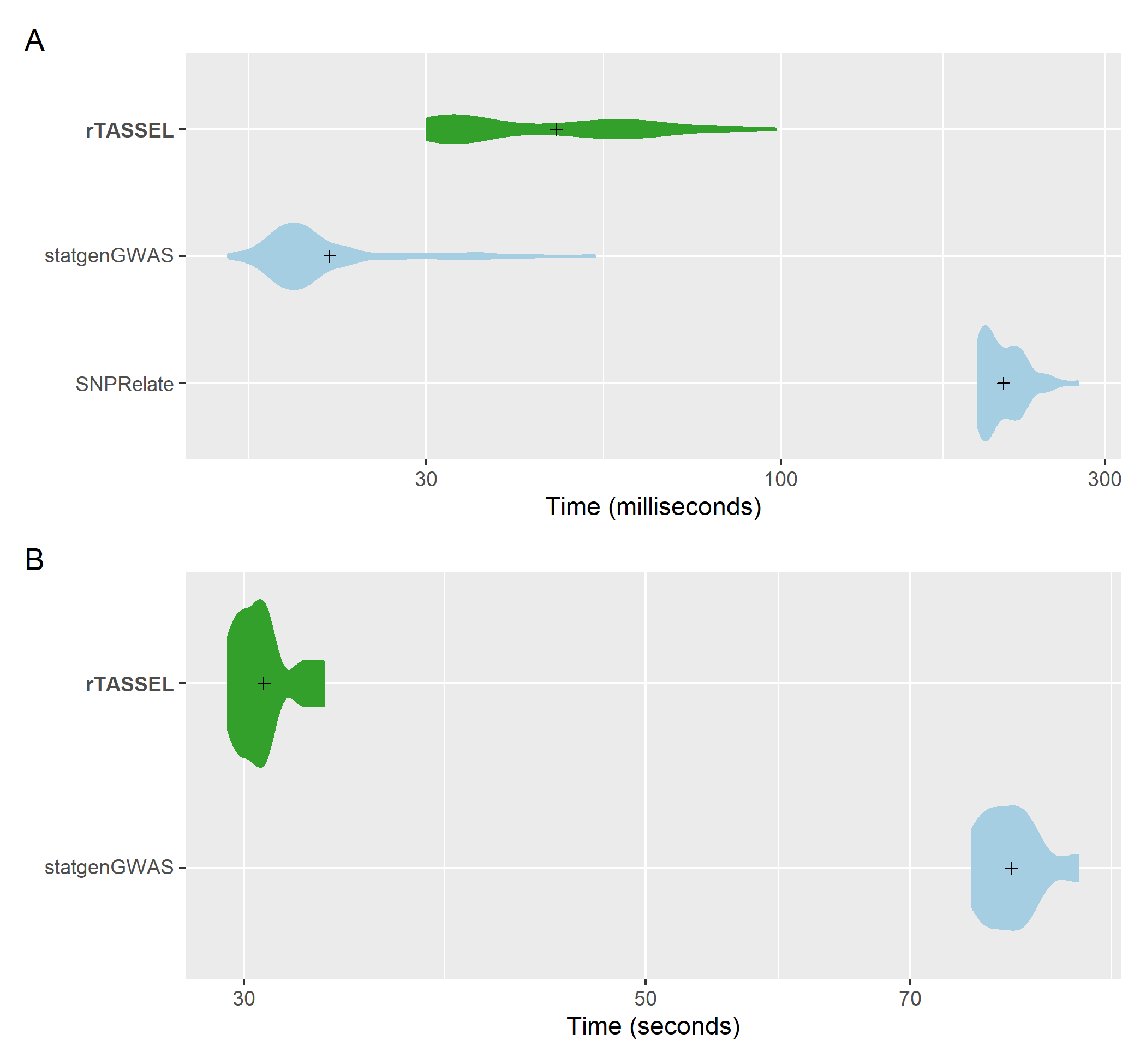
Kinship matrix (IBS) generation time
comparisons of genotypic data. A distribution of replicated
benchmark evaluations with recorded means (cross shapes) are plotted for
rTASSEL and the R packages statgenGWAS (Rossum and Kruijer, 2020) and
SNPRelate (Zheng et al., 2012). Generation times are recorded for 279
samples x 3093 variant sites (A) and 1,210 samples x 2,255,405 variant
sites (B).






